
Lengyel, Judith A., and John R. Merriam. 2001. Translation of a new genetic screening method (ectopic expression) from the research lab to a teaching lab. Dros. Inf. Serv. 84: 218-224.
|
|
|
|||
|
|
||||
Translation of a new genetic screening method (ectopic expression) from the research lab to a teaching lab.
Lengyel,
Judith A., and John R. Merriam. Department of Molecular, Cell, and Developmental Biology, UCLA, Los Angeles,
CA 90095-1606; jlengyel@ucla.edu; jmerriam@mcdb.ucla.edu
Overview
With the avalanche of genomic sequence data that is becoming available, there is a great need for undergraduates to be exposed to current techniques by which model organisms can be used to characterize gene function. As described here, we have transformed a genetic screening project currently being carried out in both our laboratories into an exercise that can be productively engaged in by undergraduates.
Drosophila has long been used to illustrate basic genetic processes
and concepts such as recombination, sex linkage, dominant versus recessive,
etc. Here, we describe a screen
based on ectopic expression leading to phenotypic changes. This is an appropriate project for undergraduates
who have previously had an introductory genetics class. We do this with up to 30 students, meeting
twice weekly for three hours per session, over the course of a 10 week quarter.
The exercise introduces students to the concepts of mutagenesis (without
requiring the use of chemicals or irradiation), mobile elements (transposons),
regulation of gene expression, and genetic engineering.
Further, it provides hands-on experiences with balancer chromosomes,
dominant and recessive alleles, segregation, and sex linkage. Finally, in generating and characterizing their own mutants,
the students can experience the scientist's thrill of finding something that
no one has found before.
Introduction
to ectopic expression screens
Ectopic expression screening has been developed elegantly by Rorth (1996). The rationale for this type of screen is that expression of a wild type copy of a gene in a pattern different from normal will give a phenotype. Many of the transcription factors and cell signaling molecules that control development are expressed in highly spatially localized patterns; the disruption of a particular pattern by forced ectopic expression is therefore expected to lead to a phenotype. Also, certain genes that function redundantly might have no identifiable loss-of-function phenotype, but might, if expressed ectopically, give a phenotype. These ideas have been validated by results of recent ectopic expression screens (Rorth et al., 1998; Toba et al., 1999; Duchek et al., 2001).
The GAL4 UAS system, based on a yeast transcription factor (GAL4) and
its DNA binding site (UAS) was developed by Brand and Perrimon (1993) to allow
ectopic expression in Drosophila. Brand and Perrimon placed expression of
GAL4 under control of a variety of temporal- and spatial-specific promoters;
further, they showed that a specific, known gene can be brought under GAL4
control by cloning it adjacent to the UAS promoter element and transforming
it back into the Drosophila
germline.
In the ectopic
expression screen described here, random, unselected genes are brought under
GAL4 control by use of a specially constructed P element. This P element carries, at one of its ends, an oligomerized
UAS (the GAL4 binding site) plus a basal promoter oriented outward (toward
the genomic DNA where it inserts). In
the presence of GAL4, genomic DNA adjacent to the element (which often inserts
at the 5' end of genes) is transcribed.
As shown in Figure 1 and in the exercise described here, the P{UAS} element is marked with y+ rather than w+; this allows the student to follow both
the P{y+ UAS} responder
and the P{w+ Gal4} driver.
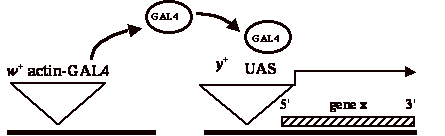 |
| Figure 1. Insertion of
P{y+ UAS} element at 5' end of
gene x brings it under GAL4 control. |
In cross I, females carrying the P{y+ UAS} construct on the X chromosome are crossed en masse to males carrying a Ki-marked chromosome that constitutively provides the P element transposase. The progeny of this cross are dysgenic, i.e., the P element is mobilized in their germ cells.
In cross II, the dysgenic male progeny from cross I are crossed individually to y w females. The progeny of this cross are screened to identify y+ w males. These y+ w males are referred to as "exceptional" males, as they carry a P element that has transposed from the X chromosome to an autosome.
In cross III, the non-Ki (so as to avoid further transposition events) exceptional males (y+ w) from cross II are crossed to females that are heterozygous for the GAL4 driver line of choice. For simplicity, we have used the actin-Gal4 driver, which is expressed globally, and thus is most likely to give a readily identified phenotype (lethality or semi-lethality). If the P element in the exceptional male has inserted in the indicated orientation at the 5' end of transcription unit "x", then any cell that expresses GAL4 will also express gene x (see Figure 1). Since the actin-Gal4 driver construct is marked with w+, a fly carrying both the P{y+ UAS} and actin-Gal4 will be both y+ and w+. We identify phenotypic effect of the actin-Gal4 driven ectopic expression by screening the progeny of cross III for absence (or reduction by at least 50%) of the y+ w+ class.
Cross IV The y+
w progeny from cross III are crossed to each other to generate
a permanent line; these progeny can also be used in further crosses to other
tissue-specific Gal4 drivers to screen for tissue-specific effects of ectopic
expression. We have found the GMR driver to be very useful, as it is eye-specific,
allowing easy characterization of phenotype in the adult. The choice of the tissue-specific driver is, of course, up
to the instructor. Many different GAL4 drivers are available through Flybase,
and some also carry a UAS-GFP construct on the same chromosome, allowing analysis
of internal phenotypes in the embryo and larva. Thus it is possible to identify
genes that, when ectopically expressed, affect develop of a particular tissue
of interest. Characterization of such insert lines, as described below, might
perhaps be part of a subsequent research project for more advanced/dedicated
students.
Expected
Results
|
Table 2. Details of
Crosses
|
|
*These light weeks can, at the instructor's discretion, be used for
additional exercises; we usually insert a meiotic mapping exercise. |
In our experience,
a class of 30 students, setting up 50 cross II's per student, can generate
approximately 600 exceptional males. Of these exceptional males, about 25%, or 150, are lethal in
crosses to the actin-Gal4 driver. Of
the lines lethal in combination with the actin-Gal4 driver, approximately
15% give a strong eye phenotype (or lethality) in combination with the eye-specific
GMR driver.
Further
Experiments
While these are
beyond the scope of a 10 week exercise, we include subsequent experiments
that could be done by the more advanced students in subsequent quarters. This
makes students aware of what one might do with an interesting insertion line
that they identify in their screen.
Mapping insert
molecularly. The insertion position of the P{y+
UAS} can be mapped to the nucleotide level,
based on the fact that the complete sequences of both the P element and the
Drosophila genome are known. DNA of the insertion line is extracted, cut with a variety
of different restriction enzymes that are know to cut at specific places in
the P element, and then ligated to generate circles containing P element DNA
and genomic DNA adjacent to the P element insert; inverse PCR is then performed
using primers directed outward from the P element into the genomic DNA (Sullivan
et al., 2000). The PCR product is then sequenced, and
the non-P element DNA sequence (which should be Drosophila genomic DNA) is BLASTed against the Drosophila genome sequence (on BDGP: http://www.fruitfly.org/). When a hit is found (i.e., the transcription unit "x" is identified),
the P element insert is thus considered mapped (this should be confirmed,
however, by other techniques).
Characterizing
gene expression. This can be determined by in situ hybridization to whole mount embryos or hand-dissected
imaginal discs. A digoxygenin
labeled DNA or antisense RNA probe is usually used. In addition to examining expression during
normal development, one will also want to determine if gene "x"
is indeed expressed throughout the tissue when driven by the Gal4 driver used
in the screen.
Making mutants
in gene "x". To understand the function of gene x, we
want to know what happens when the fly lacks the normal function of gene x.
Having a P element insert near x is not the same as having a loss-of-function
mutation in the gene itself. If
gene x is novel, and there is no known mutation in it, null mutations can
be generated by excision of the P element, which should generate small deletions
that extend into the gene.
Notes for
the student:
Genetic
notation in Drosophila
There are a number of useful reference books on Drosophila genetics (Ashburner, 1989; Lindsley and Zimm, 1992; Roberts, 1986; Greenspan, 1997). We provide here a brief introduction that should give you enough information to get started.
All Drosophila genes are written in italics and the more recent convention is for the gene to be abbreviated by three letters (e.g., dpp for decapentaplegic). Each gene abbreviation is unique. If the first allele(s) found are dominant, then the first letter of the gene name is capitalized (e.g., Cy). If the first alleles isolated are recessive, the gene is written as lower case. Note that the same gene can have both recessive and dominant alleles, so this early convention, although still used, has its problems. An exception to the rule that genes are abbreviated by three letters is that genes discovered more than 15 or 20 years ago may have a one or two letter abbreviation (e.g., y, w, f, Cy). The current convention is that different alleles are denoted by superscript numeral starting with 1, e.g., byn1, byn2, etc.; many alleles are referred to by their original notations, however, for which there is very little uniformity, e.g., w1118.
There are four chromosomes in the Drosophila haploid genome. The first chromosome is the X, which pairs with the Y; these are referred to as the sex chromosomes. There are also chromosomes 2, 3 and 4; these are referred to as the autosomes. The fourth chromosome is so small that, unless you are working on one of the few genes that have been mapped to the fourth, it can be ignored. The second and third chromosomes are divided into left and right halves by a centromere; these halves are referred to as 2L, 2R, 3L, and 3R.
Sex in Drosophila is determined by the ratio of the number of X chromosomes to the number of autosomes. Females are determined by the XX and males by the XY genotype. Only a few genes controlling male fertility map to the Y chromosome; therefore, we can ignore it.
A Drosophila stock is described
only by the mutant genes that it carries; wild type genes are not listed in
the genotype. Mutant genes on the same chromosome are written in the same
order in which they map to the chromosome, from left to right, and separated
by a space, e.g., y
w f. If there
is no designation of the genotype of the homologous chromosome (remember that
there are two homologs for each chromosome, one from the mother and one from
the father), it can be assumed that the mutant chromosome is homozygous.
Obviously this is only possible if none of the mutant alleles on the
mutant chromosome is homozygous lethal. The y w f
chromosome is, for example, homozygous viable.
Balancer
chromosomes
Balancer chromosomes are one of the unique tools of Drosophila genetics and a key reason why many types of powerful genetic screens are possible. Balancer chromosomes make it possible to generate and maintain a “balanced lethal line" (lethal is abbreviated l; standard usage is for it to be separated from the homologous balancer chromosome with a "/", indicating allelic segregation). The characteristics of a balanced lethal line are that only the genotype l/Balancer is viable, while Balancer/Balancer and l/l are dead. Thus each mating of the parental l/Balancer stock regenerates itself (while producing l/l progeny whose phenotype can be examined) and maintains the lethal allele.
There are Balancer chromosomes for the X, the second and the third chromosome. Each balancer has three essential features:
a.) a lethal mutation. This is essential for keeping a balanced lethal line, since it means that Balancer/Balancer is homozygous lethal (or female sterile, for X chromosome balancers).
b.) sufficient inversions to prevent crossing over. This isolates a mutant allele of interest on a particular chromosome with whatever markers are associated with it; also it keeps the mutant allele of interest separate from the lethal mutation on the Balancer that is necessary to maintain the balanced lethal line.
c.) a dominant mutation that allows the Balancer to be recognized. The most commonly used marker on the X chromosome is Bar eyes (kidney shaped eyes as heterozygotes, narrow, slit-shaped eyes in homozygotes). The most commonly used marker on the second chromosome is Cy (curly wings). The most commonly used marker on the third chromosome is Sb (stubby bristles on the notum, i.e., the dorsal part of the thorax). Although the nomenclature is not completely uniform, there is some regularity in the naming of balancer chromosomes. Thus balancers for the X are referred to as FM (for First (X) chromosome, Multiply inverted), for the second as SM (for Second etc.), and third as TM (for Third etc.). Also note that a common second chromosome balancer is CyO, which carries Cy. The most commonly used third chromosome balancer is TM3; it usually carries Sb.
Other common dominant markers are Glazed
(eye), Tft (bristles), and Sco (bristles) on the second chromosome, and Tb (tubby body) and Serrate (nicked wings) on the third chromosome.
Analyzing
progeny of the different crosses
Cross II: This will produce the
following male progeny by classic segregation:
y w/Y; Ki
∆2-3/+
y w/Y
However, because the P element can jump to
autosomes (2nd, 3rd or 4th chromosome) in the presence of the transposase
encoded by Ki ∆2-3, there is a low frequency of appearance of the following
males:
y w/Y; P{y+}
; Ki ∆2-3/+ Ki exceptional males
y w/Y; P{y+} non-Ki
exceptional males
Use only the
non-Ki exceptional males for Cross III (otherwise the P element will keep
jumping). Because the "jump"
resulting in some Y-bearing sperm carrying P{y+} from the dysgenic father can occur early (before meiosis)
in the father's germ line, many of the exceptional males from one father are
identical. To avoid ending up
with many lines that have the same insertion site, use only one exceptional
male from each cross II vial.
Cross III: The possible
progeny classes and their phenotypes are as follows:
i
y w/y w or Y; +/CyO (y
w Cy)
ii
y w/y w or Y; +/CyO; P{y+ UAS} (y+
w Cy)
iii
y w ; P{w+ actin-Gal4} / + (y
w+)
iv y w ; P{w+ actin-Gal4} / +; P{y+ UAS} (y+ w+)
If the P{y+ UAS} element of a particular male has inserted near a gene,
and the ectopic expression of this gene driven by the actin-Gal4 driver is
deleterious, then there will be an absence (or reduced number—less than
50% of expected) of the class iv (y+ w+) progeny. For
any exceptional male giving this result, you can map the insert to either
chromosome 2 or chromosome 3 by crossing brothers and sisters (sib mating)
the class ii (y+ w Cy) progeny. You should use Punnett squares to work out for yourself the
following:
1. If the P{y+ UAS}
maps to the 2nd chromosome, then all of the progeny of the sib mating will
be y+. If the
insertion is homozygous viable, one-third of these progeny will be straight
winged).
2. If the P{y+ UAS}
maps to the 3rd chromosome, then the progeny of the sib mating will be both
y and y+, and Cy
and straight winged, i.e., there
will be four progeny classes.
References: Ashburner, M., 1989,
Drosophila, A Laboratory
Handbook. Cold Spring Harbor Laboratory
Press, Cold Spring Harbor, N.Y.; Brand, A.H., and N. Perrimon 1993,
Development 118: 401-415; Duchek,
P., K. Somogyi, G. Jekely, S. Beccari, and P. Rorth
2001, Cell 107: 17-26; Greenspan,
R.J., 1997, Fly Pushing:
The Theory and Practice of Drosophila
Genetics. Cold Spring Harbor Laboratory
Press, Cold Spring Harbor, N.Y.; Lindsley,
D.L., and G.G. Zimm 1992, The Genome of Drosophila melanogaster, Academic Press, New York;
Roberts, D.B., 1986, Drosophila: A Practical Approach, IRL Press, Washington, D.C.; Rorth, P. 1996, Proc. Natl. Acad. Sci. USA 93: 12418-12422;
Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm,
G.M. Rubin, K. Weigmann, M. Milan, V. Benes, W. Ansorge, and S.M. Cohen
1998, Development 125: 1049-1057; Sullivan, W., M. Ashburner, and R.S. Hawley 2000, Drosophila Protocols, Cold Spring
Harbor Laboratory Press, Cold Spring Harbor, N.Y.; Toba, G., T. Ohsako, N. Miyata, T. Ohtsuka,
K.-H. Seong, and T. Aigaki 1999, Genetics 151: 725-737.