
McAllister, Bryant F. 2001. Genetic analysis of sex-chromosome arrangement in Drosophila americana: a laboratory exercise for undergraduate or advanced placement students. Dros. Inf. Serv. 84: 227-234.
|
|
|
|||
|
|
||||
Genetic analysis of sex-chromosome arrangement in Drosophila americana: a laboratory exercise for undergraduate or advanced placement students.
McAllister, Bryant F. Department of Biology, University of Texas at Arlington, Arlington, TX 76019.
This paper describes a crossing design for determining the arrangement
of the X and 4th chromosomes in Drosophila americana. Implementation
of this crossing design in a teaching laboratory with undergraduate or advanced
placement students provides a clear demonstration of sex-linked vs. autosomal
inheritance and the connection between linkage relationship and chromosomal
arrangement. Analyses can be
performed on newly collected material, or on established stocks available
from the Tucson Stock Center (University of Arizona) or the author.
The procedure requires minimal setup, takes about 10 weeks to complete,
and generates easily interpretable results.
In this description of the procedure, an example is provided where
four undergraduate students analyzed a sample (OR01) of D. americana collected near Toledo, Ohio. An electronic copy of the procedure is
available from the author upon request (bryantm@uta.edu).
Background:
Linkage relationships among loci are a product of the underlying organization of the genome into chromosomes. This was recognized very early in the history of Drosophila genetics, and provided a basis for the chromosome theory of inheritance. Change in genome organization through chromosomal rearrangement produces a corresponding change in linkage relationships. Analyses of chromosomal rearrangements and their affects on linkage are the primary method for associating physical and linkage maps in most organisms. A chromosomal rearrangement consisting of an autosome/sex chromosome translocation in Drosophila americana provides an exemplary system for demonstrating the connection between chromosomal arrangement and linkage relationship. A derived fusion of chromosome 4 with chromosome X exists in some populations of D. americana (Throckmorton 1982). In male flies with the X-4 fusion, loci on the 4th chromosome exhibit complete sex linkage as a consequence of the absence of crossing over (Figure 1). In male flies where the X is not fused to a 4th chromosome, loci on the 4th chromosome segregate as normal autosomes (Figure 1). Linkage relationship of the 4th chromosome relative to gender provides an easy method of diagnosing this chromosomal arrangement (Stalker, 1940). Cytological analysis of mitotic chromosomes can also be performed as a primary method of diagnosing this chromosomal arrangement or as a comparative exercise (Hughes, 1939).
The X-4 fusion has a distinct distribution throughout the United States, where it is observed at high frequencies in northern latitudes and at low frequencies in southern latitudes. Frequencies of the alternative arrangements are tightly correlated with latitude (McAllister, 2002). Both arrangements, fused X and 4th chromosomes and unfused X and 4th chromosomes, exist in populations that occur over a wide geographic region of intermediate latitude, probably between the 33rd and 43rd parallels. Longitudinal variation in frequencies of these arrangements has yet to be quantified. Historically, the two different chromosomal forms have been taxonomically subdivided as D. americana, containing the X-4 fusion, and D. texana, containing autosomal 4th chromosomes. Often the taxonomic designations are as subspecies of D. americana. Absence of genetic differentiation between the two chromosomal forms and widespread polymorphism for the chromosomal arrangements indicates that taxonomic subdivision is unwarranted (McAllister, 2002). The arrangements represent an intraspecific chromosomal polymorphism, where each arrangement presumably contains associated genetic variation that is distributed over this geographic gradient by natural selection.
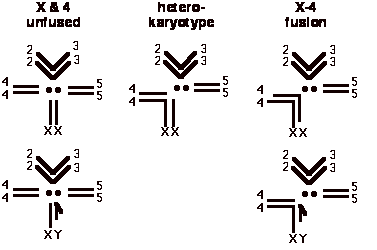 |
| Figure 1. Chromosomal organization in Drosophila americana. Notice the structure of the 4th chromosomal pair as autosomes in males when not fused to the X, and the sex linkage of this chromosomal pair when the X-4 fusion is present in males. In geographic regions where chromosomal arrangement is polymorphic, all five karyotypes exist within the population. The objective of the laboratory exercise is to determine the condition (fused or unfused) of each X chromosome within a sample of females. |
Collection and analysis of samples derived from natural populations is feasible and provides an "open-ended" laboratory experience that can involve several primary exercises, including this linkage analysis. Populations of D. americana exist in the central and eastern United States, occurring in riparian habitats. Adults can be collected throughout the spring, summer and fall in the mornings and evenings from baits of fermented banana that are placed among dense stands of sandbar or black willow near the margins of marshes, rivers or lakes (Blight and Romano, 1953). Generally, collections yield few specimens of D. americana relative to other species of Drosophila; therefore, morphological identification is necessary. Strickberger (1962) contains a full morphological key for identifying Drosophila species collected in the United States. Key features of D. americana are the large size, divergent anterior scutellar bristles, clouded crossveins, and solid black coloration on the dorsal surface of the abdomen. This species effectively lacks a mid-dorsal line (delineated by the absence of dark pigment). Collections of greater than 50 individuals are usually obtained over about three days of morning and evening collecting in the appropriate habitat.
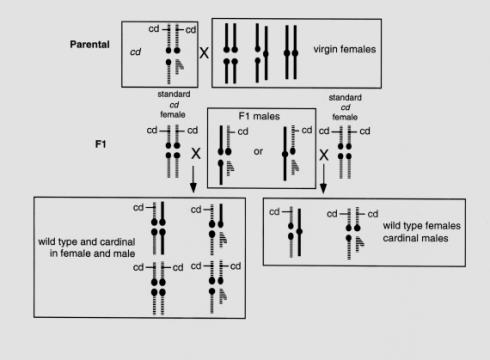 |
| Figure 2. Crossing design for analysis of X and 4th chromosomal arrangement in Drosophila americana. Virgin females are crossed to males that are homozygous mutant at the cardinal (cd) locus on the 4th chromosome, and F1 males are backcrossed to the cd stock. Segregation of cd in the F2 progeny as an autosomal or as a sex-linked locus indicates the chromosomal arrangement. |
|
Linkage analysis of the 4th chromosome relative to gender provides a simple method for analyzing chromosomal arrangement in D. americana. The procedure involves crossing a wild-type female, either recently collected and sperm depleted or from an iso-female line, to males from a standard mutant stock that is homozygous for the recessive cardinal (cd) mutation, which is located on chromosome 4. Several strains of D. virilis in the Tucson Stock Center are homozygous cd and these are interfertile with D. americana. The author has a line of cd that was obtained by crossing Stock Center strain #1051.46 with a line of D. americana and subsequently inbreeding, and this line produces larger progenies when crossed with wild-type D. americana. Homozygous cd flies have bright red eyes that are clearly distinguishable from the deep red eyes in wild-type flies. The resulting F1 males are individually backcrossed to the cd stock to determine if chromosomal pair 4 is sex-linked, or not. Each of the 4th chromosomes in the F1 male carries a specific allele, the wild 4th (maternal) contains a wild-type allele and the lab-4th (paternal) contains the mutant allele. Phenotypic results in backcross progeny are used to determine if the wild 4th chromosome is fused to the X and transmitted only to females, or not fused to the X and transmitted equally to females and males (Table 1). An overview of the crossing design is presented in Figure 2.
Using appropriate sam-pling, this crossing design can be used to determine the arrangement (fused to 4, or not fused to 4) of X chromosomes within a sperm-depleted wild-caught female or an iso-female line based on the segregation of the cd locus in F1 males. The wild-caught females have two X chromosomes, and implementa-tion of the appropriate experimental design provides data on the arrangement of both chromosomes. Iso-female lines are established by allowing a single wild-caught female to lay eggs, and the progeny are subsequently maintained each month. At least three X chromosomes are present in each line; two from the wild-caught female, and one from her mate (if she mated to more than one male, there will be more than 3 X chromosomes in the line). A detailed protocol for the analysis of iso-female lines is provided as an Appendix. By crossing either type of wild-type females to a standard cd stock and following the segregation of cd in the F2 progeny of backcrossed F1 males, arrangement of the X chromosome in each F1 male can be easily inferred (Table 2).
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Over the period of May 20 to 22, 2001, a sample of D. americana was collected at the Ottawa National Wildlife Refuge (GPS: 41˚ 36.68 N, 83˚ 12.13 W) in Oak Harbor, Ohio. Bait was prepared by mashing ripe bananas with baker's yeast and fermenting for several days. About three tablespoons of fermented banana were placed in plastic drinking cups, which were hung with string from tree limbs in marshy areas containing a high density of sandbar willows. Flies were collected from the baits in the mornings and evenings, and immediately after each collection the flies were anesthetized with CO2 in order to identify D. americana. Individual females were separated into vials containing standard cornmeal medium sprinkled with active dry yeast, and males were isolated into groups. A total of 50 females and 34 males were collected and transported to the laboratory. Of the 50 females collected, 21 produced progeny that were used to establish iso-female lines. The wild-caught females were serially transferred in the laboratory until they failed to produce offspring. The frequencies of the two chromosomal arrangements were obtained for this sample of females using the crossing design presented.
Both wild-caught females and iso-female lines were used to examine the chromosomal arrangements in this sample. The iso-female lines were assayed according to the protocol described in the Appendix. A different sampling approach was used for wild-caught females. For these females, linkage relationship was examined for multiple F1 males in order to infer the arrangement of both X chromosomes in each female. About 12 F1 males were individually backcrossed for each wild-caught female. In calculating the frequencies of the alternative chromosomal arrangements in the sample, results for at least six F1 males were necessary to infer the arrangements of both X chromosomes in the wild-caught female. When results were obtained from less than six F1 males, the status of a single X chromosome was randomly inferred for the female (even if the female was clearly a heterozygote).
The analysis of wild-caught females provides unambiguous assignment of chromosomal arrangements when linkage relationships are examined in a sufficient number of F1 males. Table 3 presents the number of X-4 fusion chromosomes and unfused X chromosomes that were observed in the analysis of the F2 progeny of 40 different wild-caught females. Of these 40 females, 19 were homozygous for X-4 fused chromosomes, 5 were heterozygous and 2 were homozygous for unfused X chromosomes. These frequencies of genotypes are not inconsistent with Hardy-Weinberg equilibrium. In the 14 other females, an insufficient number (less than 6) of F1 males produced F2 progeny, so the status of only a single X chromosome in the female was inferred.
Inference of chromosomal arrangements for multiple chromosomes in an iso-female line is confounded by potential differences in frequencies of the chromosomes in the line, presence of more than three X chromosomes in the line, and inference of proportions for the three chromosomes in the line based on a small sample. Even with these issues, the data obtained from the analysis of iso-female lines derived from this sample of D. americana is consistent with the data obtained from the analysis of wild-caught females (Table 3). Although a difference of 4.2% was observed for the estimates of the frequency of the X-4 fusion, this difference is not statistically significant. The analysis of iso-female lines is a reliable method of inferring the frequencies of the alternative chromosomal arrangements in D. americana, and its implementation with students in a teaching laboratory is more feasible than analyses of wild-caught females.
|
Analysis of this particular sample collected near Toledo, Ohio was based upon both wild-caught females and iso-female lines, and the data obtained from these two methods are not entirely independent in respect to chromosomes sampled from this population. This is due to the fact that in some cases, an individual female's X chromosomes were analyzed and the iso-female line she established was also analyzed. Table 3 presents the combined data, accounting for instances of replication, for the number of X-4 fusion and unfused X chromosomes that were independently sampled and the estimated frequency of X-4 fusion chromosomes at this locality. The estimate of 84.3% X-4 fusion chromosomes at this locality is lower than would be predicted (greater than 100%) based on the regression of this chromosomal arrangement relative to latitude in the Mississippi River Valley (McAllister 2002). The high frequency of unfused X chromosomes in this population suggests that there is a longitudinal pattern in the distribution of these alternative chromosomal arrangements in D. americana. Estimates of the frequencies of the arrangements from many samples of D. americana from throughout the central and eastern United States are needed to effectively examine this pattern.
This procedure for analyzing the arrangement of the X and 4th chromosomes in D. americana is an effective exercise to incorporate into a laboratory course. This exercise illustrates fundamental genetic concepts concerning genome organization. In a laboratory course, coupling this linkage analysis with other experimental approaches provides a comprehensive exposure to experimental genetics. Chromosomal arrangements within iso-female lines can be examined directly by examining mitotic chromosomes prepared from larval ganglia. Molecular markers on the 4th chromosome could also be used to infer chromosomal arrangements through linkage. Both RFLP markers (McAllister, 2002) and microsatellite markers (Schlötterer, 2000) are available for these purposes. The utilization of crossing, chromosomal, and molecular analyses to examine chromosomal organization in D. americana provides a broad exposure to different experimental methods.
Acknowledgments: The efforts of Mathew Spinks, Weldon Turner, Phillipe York, and Cory Zablonsky in the analyses of the OR01 sample are greatly appreciated. I thank Ron Huffman of the Ottawa NWR for assistance in using the refuge for collecting flies. This project was supported by DEB-0075295 from NSF.
References: Blight, W.C., and A. Romano 1953, American Naturalist 87: 111-112; Hughes, R.D., 1939, Genetics 24: 811-834; McAllister, B.F., 2002, Genome 45: 13-21; Stalker, H.D., 1940, Proc. Natl. Acad. Sci. USA 26: 575-578; Schlötterer, C., 2000, Heredity 85: 610-616; Strickberger, M.W., 1962, Experiments in Genetics with Drosophila. John Wiley and Sons, Inc.; Throckmorton, L.W., 1982, In: The Genetics and Biology of Drosophila, vol. 3b. (Ashburner, M., H.L. Carson, and J.N. Thompson, jr., eds.). pp. 227-296. Academic Press, London.
APPENDIX
The following is a detailed protocol that can be used to examine the X chromosomes in an iso-female line.
Materials needed: Iso-female lines of Drosophila americana, homozygous cardinal stock of Drosophila americana, vials containing fly medium (standard cornmeal medium, or commercially prepared medium), routine supplies for fly experiments (dissecting scopes, anesthesizers, paintbrushes).
Prior to beginning the experiment, students should familiarize themselves with the separation of female and male flies. Male D. americana do not have sex combs; therefore, gender is determined by abdomen shape and external genitalia.
Protocol:
1. Obtain virgin females from a wild-type line. Clear adults from vials that you intend to collect females. Within 4-5 days of eclosion, collect and isolate females from the vials. Place females in a clean vial with food and allow them to sexually mature (5-7 days). A maximum of 20 females from each line is needed.
2. Collect males from stock bottles of a cd line. This is a mutant strain that is homozygous for the visible mutation cardinal, which is located on the 4th chromosome and yields a bright red eye color. Collect at least 2 males for each of the females that were collected in step 1, so about 40 males per iso-female line. Place males in groups in clean vials with fresh medium, and allow to reach sexual maturity (5-7 days).
3. Set up parental crosses. Sprinkle dry yeast on the surface of vials containing fresh food. In each vial, add two cd males and a single virgin female. It is easiest to set up many vials containing yeast and two cd males, and then add a single female to each vial. Label vials with the iso-female line and number each (1, 2,…). Set up 14 crosses for each iso-female line with which you are working. Store the crosses between 18-24˚C.
4. Clear parents. About a week after the crosses were set up, clear the adults from the vials and place in a morgue (dish soap and water). Larvae should be visible at this point.
5. Collect F1 males. About 3-4 weeks from the date that a cross was initiated, adults will begin to emerge in the vials. Collect about 5-10 males from each of the vials and place them in a vial containing fresh medium. Note: All F1 flies should have wild-type eyes.
6. Collect virgin cd females. At about the same time as the flies from the crosses are emerging, clear the adults from bottles of the cd stock and collect females within 3-4 days. About 140 cd females are needed for each iso-female line that is being analyzed. Place groups of about 20 females in vials containing fresh food.
7. Set up F1 crosses. After the collected flies have aged at least one week, set up the backcrosses with the F1 males. Place about 4 cd females in a vial containing fresh food sprinkled with yeast. Add a single F1 male to each of these vials. Cross at least two F1 males from each of the vials containing parental crosses. Assuming that all 14 parental females produced at least two F1 males, 28 crosses will be set. Label each of these vials with the name of the original line, the number on the vial of the parental cross, and give each F1 male a unique ID (such as A and B). Example, FP99.2-2A, which means line FP99.2, parental cross 2, F1 male A.
8. Clear parents. About a week after the crosses were set up, clear the adults from the vials and place in a morgue.
9. Analyze F2 progeny. The backcross progeny will emerge within about 3-4 weeks after the crosses were initiated. After about 20 progeny have emerged in a vial, record the sex and eye color of each fly. For each pair of brothers that were crossed, it is unnecessary to record the results from both (two crosses were set to ensure that results were obtained for the parental female). It may be necessary to count flies on at least two occasions, because flies emerge from some vials slower than others. For each family of backcross progeny, determine the organization of the wild X and 4th chromosome in their father (a single F1 male). If all the male progeny have wild-type eyes, and all the female progeny have bright-red eyes, the X and 4th chromosomes were fused in their father. If gender and eye color are independent, the X and 4th chromosomes in their father were not fused.
Data analysis:
Data from each family of F2 progeny can be examined statistically by testing the hypothesis of independent assortment of gender and eye color using a 2 x 2 test of independence and the chi-square or G statistic. Sufficient data are usually obtained for a family of F2 progeny when about 15 flies are counted, and either all progeny fall into the two linked phenotypes or all four phenotypic categories are represented. Two examples of F2 data are provided in Table 1. For each iso-female line, assessment of the status of at least nine X chromosomes in the line provides a high probability (>95%) of assaying all of the chromosomes present in the line, assuming only three X chromosomes are present at equal frequency. As long as results are obtained for one F1 male (at least two should have been set) for each of 9 parental females (at least 14 should have been set), appropriate numbers will be obtained.
The alternative results for a given iso-female line (counting a single F1 male for each female from the line and having data for at least nine F1 males) are as follows:
3 fused X-4 chromosomes: All F1 males from a line transmit cd in a sex-linked pattern.
2 fused X-4, 1 unfused X: Greater than half of the F1 males from a line transmit cd in a sex-linked pattern, less than half are autosomal.
1 fused X-4, 2 unfused X: Less than half of the F1 males from a line transmit cd in a sex-linked pattern, greater than half are autosomal.
3 unfused X chromosomes: All the F1 males from a line transmit cd in an autosomal pattern.
Using this basis, the frequency of the alternative chromosomal types in a sample can be calculated for an entire class that has examined multiple iso-female lines.