
Gu, Sheng, Yi Gong, Matthew Smith, and R.C. Woodruff. 2002. The dominance value of the Sb allele in Drosophila melanogaster. Dros. Inf. Serv. 85: 143-149.
|
|
|
|||
|
|
||||
The dominance value of the Sb allele in Drosophila melanogaster.
Gu, Sheng, Yi Gong, Matthew Smith, and R.C. Woodruff. Department of Biological Sciences, Bowling Green State University, Bowling Green, OH.
Introduction
The genetic material of all organisms, including humans, seems remarkably stable. For example, most humans seem to develop from zygote to adult with few major morphological mistakes. There are few people with two heads or three arms. Yet this assumption of genetic stability is an illusion. For example, every cell of a human undergoes hundreds of nucleotide changes and at least ten double-stand breaks in DNA occur each day (Bernstein and Bernstein, 1991). The reason we survive this genetic insult is that most of this genetic damage is corrected by DNA repair mechanisms. However, not all of these mutations are corrected. For example, it has been estimated that at least 100 nucleotide changes occur in each human gamete, each gamete has from two to four new deleterious mutations, and each human has one or more recessive lethal mutations (Crow, 2001; Eyre-Walker and Keightley, 1999; Kondrashov, 2001). These deleterious and lethal mutations can have an important effect on our health and fitness. However, the influence of these mutations depends on their selection values (how much does each mutation reduce health and fitness) and on their genetics (are they recessive or dominant).
It is usually assumed that recessive lethal mutations also have some influence on the health and fitness of individuals when these mutations are present as heterozygotes, i.e., the individual without this mutation (+/+) is assumed to be more healthy and fit (produces more children, for example) than the individual that is heterozygous (+/l) for this mutation. This reduction in health and fitness in the heterozygotes would be due to some amount of dominance for the lethal mutation. What is this level of dominance (called h, the dominance value) and how does one determine its value?
One way to visualize the influence of a mutation on fitness is to use the following standard population genetic model, where s = selection coefficient, h = dominance value, maximum fitness = 1, and the gene is assumed to have two alleles + and l.
| Genotypes: | + / + | + / l | l / l |
| Fitness: | 1 | 1 - hs | 1 - s |
| With s = 1 (lethal) | 1 | 1 - h | 0 |
Therefore, the fitness of the +/l heterozygous individual is dependent on the dominance of the l allele. If the l allele has a dominance of 1, the heterozygotes have a fitness of zero (die early or produce no progeny), whereas a dominance value of less than one will reduce fitness by that amount; for example, an h value of 0.4 would give a fitness of 1 - 0.4 = 0.6 for +/l individuals. In this example, the individuals would be only 60% as fit as the +/+ individuals or produce 40 percent fewer offspring.
Experiment
Design
|
Figure 1. Mating scheme to collect Sb/+ females and Sb/+ males after six generations. |
The following experiment is designed to calculate the dominance value (h) for the third-chromosome Stubble (Sb) mutation of Drosophila melanogaster. With the data collected from this experiment, h values can be roughly estimated through the formula provided by Chung (1967).
Two stocks are needed for this experiment: In(3LR)Ubx101/Sb and the wild type Canton-S. Stubble (Sb) is a dominant visible mutation causing short bristles and is also a recessive lethal (i.e., Sb/Sb flies die as embryos). (For details, see Lindsley and Zimm, 1992; or http:// flybase.bio.indiana.edu). The following crosses were performed to collect virgin Sb/+ females and Sb/+ males.
To get a precise estimation of
h for the Sb mutation, we set up two population cages (Cage A and
Cage B), each containing 100 virgin Sb/+ females and 100 Sb/+ males.
Therefore, the starting frequency of the Sb allele was 0.5.
Specifications and Details
Cage Size: 11 ´ 8 ´ 4.5 inches, with 15 holes on the base for15 vials
Vial Size: height 3.75 inches, diameter 0.88 inch
Food Change: Replacement of four new vials containing food every week for 15 weeks (six generations)
Temperature: 24~26ºC
Average generation interval: 17 days
Materials and Method:
t = (1-h)/h
* ln{[h+qt(1-2h)]/[h+q0(1-2h)]} +1/h * ln(q0/qt)
(I)
| Genotype | +/+ | Sb/+ | Sb/Sb |
| Fitness: | 1 | 1 - hs | 1 - s |
| Since Sb/Sb is lethal, s = 1 and: |
|||
| Fitness | 1 | 1 - h | 0 |
| Frequency | p2 | 2pq | q2 |
since p =
1 - q,
Δq = [q(1-h) -q(1-q) -2q2(1-h)] / [1-q+2q(1-h)] = - [q(q+h-2qh)] / (1+q-2qh)
Since the
population is changing continuously, treat Δq as approximately dq/dt.
Therefore,
dq/dt = - [q(q+h-2qh)] / (1+q-2qh)
dt = (1+q-2qh) / [-q(q+h-2qh)] dq
Let (1+q-2qh) / [-q(q+h-2qh)] = M/-q + N/(q+h-2qh)
Balance both sides and we get
M = 1/h
N = (2h-1)(h-1) /
h
Hence:
t = ò dt = ò (1+q-2qh) / [-q(q+h-2qh)] dq = ò M/-q dq +ò N/(q+h-2qh) dq
t = (1-h)/h
* ln{[h+qt(1-2h)]/[h+q0(1-2h)]} +1/h * ln(q0/qt)
(I)
Results
We chose the period from week 1 to week 16, covering roughly six generations (around 17 days per generation), to estimate h. Using CA-CricketGraphIII’s logarithmic curve fit, we got the two curves shown in Figure 2. They are very close and have nearly the same shape. The estimation of h was made using equation I and Mathematica 4.0 (or use Maple, Matlab, or other program) as follows:
Cage A: q0 = 0.5
qt = -0.312Log10X +0.486 = -0.312Log1016 + 0.486 = 0.11
t = 6
![]() hA »
0.12
hA »
0.12
Cage B: q0 = 0.5
qt = -0.307Log10X +0.480 = -0.307Log1016 +0.480 = 0.11
t = 6
![]() hB »
0.12
hB »
0.12
Table
1. Population cage experiments.
Week
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
Pop A
|
0.5 |
0.32 |
0.36 |
0.31 |
0.27 |
0.26 |
0.25 |
0.22 |
0.2 |
0.16 |
0.18 |
0.16 |
0.11 |
0.12 |
0.12 |
0.08 |
Pop B
|
0.5 |
0.32 |
0.34 |
0.28 |
0.26 |
0.3 |
0.25 |
0.24 |
0.22 |
0.13 |
0.17 |
0.15 |
0.11 |
0.09 |
0.11 |
0.12 |
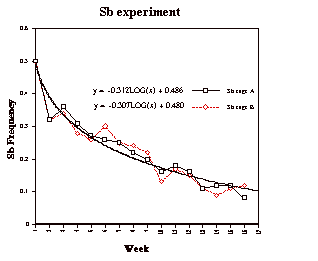
Figure 2. The change of Sb frequency in population cage A and B during 16 weeks.
Parallel Bottle Experiment
Table
2. Bottle experiment.
Generation
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
Bot
A
|
0.5 |
0.33 |
0.27 |
0.26 |
0.20 |
0.16 |
0.20 |
0.16 |
Bot
B
|
0.5 |
0.33 |
0.25 |
0.18 |
0.17 |
0.12 |
0.10 |
0.11 |
Bot
C
|
0.5 |
0.32 |
0.26 |
0.19 |
0.08 |
0.08 |
0.05 |
0.06 |
Bot
D
|
0.5 |
0.34 |
0.24 |
0.21 |
0.16 |
0.13 |
0.12 |
0.08 |
Bottle A: q0 = 0.5
qt = -0.363Log10X +0.467 = -0.363Log108 + 0.486 = 0.139
t = 7
![]() hA »
-0.026
hA »
-0.026
Bottle B: q0 = 0.5
qt = -0.445Log10X +0.474 = -0.445Log108 + 0.474 = 0.072
t = 7
![]() hB »
0.189
hB »
0.189
Bottle C: q0 = 0.5
qt = -0.518Log10X +0.489 = -0.518Log108 + 0.489 = 0.021
t = 7
![]() hC »
0.444
hC »
0.444
Bottle D: q0 = 0.5
qt = -0.449Log10X +0.481 = -0.449Log108 + 0.481 = 0.076
t = 7
![]() hD »
0.174
hD »
0.174
Discussion
The result of the population cage experiment (h = 0.12) is close to 0.16 — the h value obtained from Chung’s (1967) 23-week experiment. With more generations, the curve fit should provide a more accurate approximation of the Sb h value. However, for a laboratory exercise that can be performed by students during one semester, our bottle experiment is an appropriate design, for it lasts a shorter time (can be completed in one semester) but still gives satisfactory results.
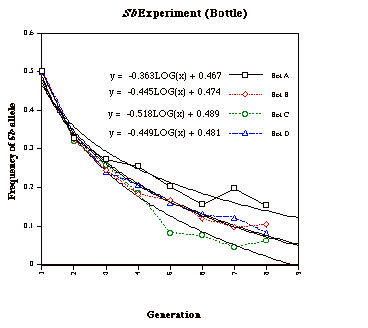
Figure 3. The
change of Sb frequency in Bottle A,
B, C and D during 8 generations.
The results of the bottle experiment show more fluctuations than those from the population experiment. In comparison with the population cage resultsand Chung’s data (1967), the result from bottle A seems unreasonably low, while that from bottle C seems high. Since we counted flies each generation andthere are eight total generations, including G0, the curve fit from only eight data points can hardly provide an accuracy as good as that of the population experiment, which had 16 data points. Although two of the bottle results seem not to be reasonable, the other two are very close to the h value obtained from Chung’s (1967) 23-week experiment. If we repeat the bottle experiment with additional bottles, we expect that the results will show a normal distribution, with most being around the normal expectation and a few outliers. Hence, compared with the population cage experiment, the bottle experiment saves labor at the cost of result accuracy. But the deviation is tolerable, and therefore it is an alternative choice for students to complete the experiment in a laboratory class during one semester. It should be noted that this technique could also be used with other dominant visible mutations that have complete penetrance and constant expression, and are also homogenous lethal, such as Plum (Pm, eye color) and brown dominant (bwD, eye color).
From the above, it is clear that an experiment of 16 weeks or seven generations is both a good experience for students to explore a basic population genetics problem (estimation of dominance) using a traditional genetics research tool — Drosophila.
In a discussion session at the end of the experiment, students might be asked to go to the National Center for Biotechnology Information web page (http://www3.ncbi.nlm.nih.gov) and identify some human traits that have a dominant phenotype and are also homozygous recessive lethals, such as achondroplasia, and the students could be asked if h, s or fitness estimations are given for their traits and why these human mutations are not eliminated with time by selection.
References: Bernstein, C., and H. Bernstein 1991,
Aging, Sex and DNA Repair. Academic Press Inc., San Diego, USA; Chung, Y.J., 1967, Genetics 57: 957-967;
Crow, J.F., 2001, Nature Rev. Genet. 1: 40-47;
Eyre-Walker, A. and P.D. Keightley 1999, Nature 397: 344-347; Kondrashov, A.S., 2001, Trends in Genetics 17: 75-77; Lindsley, D.L., and G.G. Zimm 1992, The Genome of Drosophila melanogaster. Academic Press Inc., San Diego,
USA.