
Grishaeva, T.M., S. Ya Dadashev, and Yu.F. Bogdanov. 2001. Gene CG17604 of Drosophila melanogaster predicted in silico may be the c(3)G gene. Dros. Inf. Serv. 84: 84-88.
|
|
|
|||
|
|
||||
Gene CG17604 of Drosophila melanogaster predicted in
silico may be the c(3)G gene.
Grishaeva,
T.M., S. Ya Dadashev, and Yu.F. Bogdanov. The Vavilov Institute of General Genetics,
Moscow 119991, Russia.
Among more than 80 genes controlling meiosis in Drosophila
melanogaster (Grishaeva and Bogdanov,
2000), only one gene, c(3)G, crossover suppressor on 3 of Gowen localized
in the 89 2-5 region (Nelson and Szauter, 1992), is undoubtedly involved in
the formation of the synaptonemal complex (SC) (Smith and King, 1968). The latter authors suggested that the
c(3)G gene encodes an SC protein. This
assumption was experimentally tested using molecular analysis of c(3)G.
In the abstracts of the 17th Drosophila Research Conference (1-5 September 2001, Edinburgh), R.S. Hawley and
coworkers reported that c(3)G encodes a protein that is structurally similar
to yeast Zip1p and to mammalian SCP1p, which are components of the SC (Wayson
et al., 2001). Somewhat earlier
(in August 2001) we have completed computer-aided analysis of virtual genes
localized within a broad (250 kb) interval overlapping the c(3)G localization
region and their virtual protein products. We have established that this region contains only one gene
predicted in silico whose virtual protein meets the requirements of the molecular
structure of an SC protein, namely a protein of transverse SC filaments in
D. melanogaster. This gene is
CG17604 located at position 36250-36253 kb on the NCBI molecular map. We suggest
that G17604 is most likely the c(3)G gene. A detailed account of our study will be published elsewhere
(Bogdanov et al., 2002). Our
in silico findings are in good agreement with experimental evidence of Wayson
et al. (2001) and confirm it.
Here, we present a brief description of our approach to the
problem to demonstrate that similar approach may be used for other genes coding
morphological traits (ultrastructure, for instance) in predicting their gene
products and function.
The D. melanogaster genome is sequenced in total and probable genes have been predicted in silico (Adams et al., 2000). Computer databases such as FlyBase, NCBI (National Center for Biotechnology Information), and INTERNET programs for their processing are available. Using the programs (ProtParam tool and ISREC, respectively), physicochemical properties of predicted gene products and their secondary structure can be estimated.
Our study was
aimed at analyzing in silico the predicted
products of genes from the 88Е-89В region of Bridges’ cytological
map (D. melanogaster chromosome 3R), which overlaps the region of c(3)G
localization, and comparing their secondary
structure with that of the known SC proteins of other biological species. Although SC proteins are not known in
Drosophila, these proteins were isolated and sequenced in several
mammalian species (Meuwissen et al., 1992, 1997;
Dobson et al., 1994; Liu et al., 1996) and in yeast Saccharomyces cerevisiae
(Tung and Roeder, 1998; Dong and Roeder, 2000). In view
of this, in our identification of the C(3)G protein we proceeded from the ultrastructural
characteristics of the SC in Drosophila and other organisms to those of the known SC proteins
in yeast and mammals and further to the virtual proteins whose genes were
predicted in the c(3)G gene region.
|
Table 1. The
relationship between the size of the protein molecule forming the SC
transverse filament and the SC central space width. Hs - Homo sapiens; Mm - Mus musculus;
Rn - Rattus norvegicus. For references see: Meuwissen et al. (1992, 1997) and Liu et
al. (1996) -- for SCP1; Sym and Roeder (1995) and Dong and Roeder (2000)
-- for Zip1.
|
The synaptonemal complex (SC) is a long protein structure that appears between the synaptic homologous chromosomes in the first meiotic prophase. It consists of parallel lateral elements that surround the central space containing the longitudinal central element (von Wettstein et al., 1984). The SC is rather conservative. The width of the central space of the SC is similar in different taxones: 90-120 nm in fungi, 100-120 nm in insects, about 100 nm in mammals (Westergaard and von Wettstein, 1972; von Wettstein et al., 1984). The central space is crossed by protein filaments, the SC transverse filaments. The secondary structures of the SCP1/SYN1 proteins forming the transverse filaments in mammals (rat, mouse, hamster, human), and protein Zip1 in yeast, are strikingly similar. Their central domain forms a road-shaped coiled coil whereas terminal domains have the globular structure (Liu et al., 1996; Dong and Roeder, 2000). Each transverse filament consists of two protein molecules in the oposite “head - to -head” (N-end to N-end) orientation.
At the first stage of our study we examined the relationship between the width of the SC central space in yeast and mammals and the length of the full-size SCP1 and Zip1 molecules, respectively. Next, we included in the analysis data on the yeast Zip1 protein having internal deletions and duplications of various lengths (Sym and Roeder, 1995; Tung and Roeder, 1998). This analysis was carried out on the basis of literature data (Meuwissen et al., 1992; Liu et al., 1996; Tung and Roeder, 1998) using the STATISTICA (StatSoft) software package. The coefficient of correlation between the above mentioned parameters was rather high (r = 0.85 at p < 0.001).
The correlation between the width of the central SC space and the length of the central (rod-shaped) domain of these proteins forming the coiled coil was even stronger (r = 0.90 at p = 0.001) comparatively to total protein molecules. Since the width of the SC central space in D. melanogaster is 109 nm (Carpenter, 1975), we assumed that the length of the central domain of the sought protein of transverse filaments falls between 730-970 amino acid residues at the 0.95% confidence interval (Table 1).
|
Table 2. Isoelectric
points (pI) of SC transverse filament proteins and their domains as predicted
by the use of ProtParam computer program. Hs - Homo sapiens; Mm - Mus musculus; Rn - Rattus norvegicus.
|
We have studied
in silico 78 predicted genes from the
88E6-89B2 region, which contains 250 kb (according to the NCBI database). The protein products of these genes have
not been identified earlier. It
was shown that the virtual product of only one gene can form a coiled coil
of the length that is required for two partially overlapping molecules to
correspond to the length of the transverse SC filaments in Drosophila. Only
the central 607-aa part of this virtual protein has the coiled-coil structure
whereas its N- and C-end domains are globular. This single gene is CG17604 located
on chromosome 3 at position 36250 - 36253 kb according to the NCBI molecular
map.
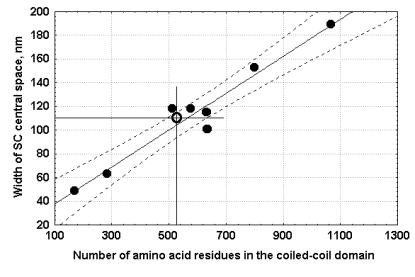 |
| Figure 1. Width of the SC central space vs. length of the coiled-coil domain of the various yeast, S.cerevisiae, Zip1 proteins of wild type and mutants with different internal deletions and duplications of various size, as calculated from Sym and Roeder (1995); and Tung and Roeder (1998). Inclined line is the line of regression. Dotted lines limit the confidence interval (P=95%). Open circle designates D. melanogaster SC central space width vs. the coiled-coil domain size of the predicted CG17604 protein. |
At the second
stage of this study we evaluated similarity between the length of the coiled-
coil part of the CG17604-encoded protein and the length of similar parts of the known proteins
of the transverse SC filaments in other organisms and then assessed the relationship
between this length and the width of the SC central space in D. melanogaster. We estimated the correlation between the width of the SC central
space in S. cerevisiae mutants
carrying internal deletions and duplications of the structural part of Zip1
and length of the corresponding coiled-coil Zip1 regions according
to Sym and Roeder (1995) and Tung and Roeder (1998).
In this case (see Figure 1) the coefficient of correlation was even
higher (r = 0.97 at p < 0.001) than that between the width of the SC central
space and the lengths of the full-size Zip1 and SCP1 molecules. We plotted the size of the central CG17604 domain and
the width of the SC central space in D. melanogaster on this regression curve (Figure 1). The size of the coiled-coil part of CG17604 corresponded perfectly
to the width of the
According to the evidence on homology of the amino-acid sequence (FlyBase), the predicted CG17604 product is homologous to various proteins, many of which can form coiled coils. These proteins include heavy myosin chain (D. melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Homo sapiens, Dugesia japonica), laminar proteins (Mus musculus), the protein of the spindle polar body NUF1 (S. cerevisiae).
Similar
homology was shown earlier for SC proteins: Zip1 in yeast (Dong and Roeder,
2000), SCP1 in rat and mouse (Meuwissen et al.,
1992; Liu et al., 1996). Thus,
the predicted CG17604 gene of
D. melanogaster is similar to
genes for SC transverse filament proteins
of other organisms also in this character. Interestingly,
the amino-acid sequences of all of the three proteins -- CG17604, SCP1/SYN1,
and Zip1 -- lack homology.
In addition, we have examined 200 predicted proteins whose genes were mapped to 12 localization regions of other meiotic mutations in D. melanogaster. It was shown that none of these 200 proteins could form coiled coils of the required length.
Using the ProtParam procedure, we estimated the isoelectric point (pI) of the known SC proteins and the found CG17604 gene product. We analyzed the total molecule and separately its domains (Table 2). In CG17604 all parameters, except the pI of the N-end domain, were similar to the parameters of Zip1 and SCP1. It is known that C-end domains of these proteins are associated with DNA of the lateral SC elements, whereas N-end domains are oriented into the SC central space, where they overlap and form the SC central element (Liu et al., 1996; Dong and Roeder, 2000). The acidic nature of the N-end CG17604 domain may reflect the specific morphology of the SC central element in Drosophila, which displays distinct striation and relatively large width of 32 nm (Carpenter, 1975a).
Thus, the discovered CG17604 gene product is similar to the known SC proteins with regard to most parameters studied. According to FlyBase, the CG17604 gene is located in the 89A7-8 region and is flanked by known genes Po and Aldox-1 to the left and by mor to the right. The c(3)G gene is localized between the same markers (Nelson and Szauter, 1992). Precise localization of predicted genes is problematic. Flybase and NCBI databases show discrepancies in their localization. Moreover, this part of 3R chromosome was shown to contain intercalary heterochromatin (Matsubayashi and Oguma, 1994) that makes it difficult to combine molecular and cytogenetic maps. Hence, we assume that the predicted CG17604 gene codes for the transverse filament protein of SC central space in D. melanogaster and is likely to coincide with the known gene c(3)G. If this is true, the c(3)G gene is located in the 89A7-8 region of Bridges’ cytological map rather than in the 89A2-5 region or even 89A2 as was reported earlier (Nelson and Szauter, 1992; FlyBase). Alternatively, if localization of c(3)G at 89A2-5 is correct, CG17604 must be localized at the same position contrary to FlyBase data. Both assumptions can be verified experimentally.
Acknowledgments: This work was supported by the Russian Foundation for Basic Research (RFBR) Grant #99-04-48182.
References: Adams,
M.D., S.E. Celniker, R.A. Holt,
et al., 2000, Science 287: 2185-2195; Bogdanov,
Yu.F., T.M. Grishaeva, and S.Ya. Dadashev 2002, The CG17604 gene of Drosophila melanogaster is a putative functional homolog of yeast Zip1 and
mammalian SCP1 (SYCP1) genes coding for synaptonemal complex proteins. Rus. J. Genet. 38, #1 (in press); Carpenter,
A.T.C., 1975, Chromosoma 51:
157-182; Dobson, M.J., R.E. Pearlman,
A. Karaiskakis, B. Spyropoulos, and P.B.Moens 1994, J.
Cell. Sci. 107: 2749-2760; Dong,
H., and G.S. Roeder 2000, J.
Cell Biol. 148: 417-426; FlyBase
Drosophila database (http://flybase.bio.indiana.edu/); Grishaeva, T.M., and Yu.F. Bogdanov 2000,
Rus. J. Genet. 36: 1301-1321; Liu,
J.G., L. Yuan, E. Brundell, B. Björkroth, B. Daneholt, and C. Höög
1996, Exptl. Cell Res. 226: 11-19; Matsubayashi, H., and Y. Oguma 1994, Jpn J. Genet. 69: 790;
Meuwissen, R.L.J., H.H. Offenberg, A.J.J.
Dietrich, A. Riesewijk, M. van Iersel, and C. Heiting 1992, EMBO J. 11: 5091-5100; Meuwissen, R.L.J., I. Meerts, J.M.N. Hoovers,
N.J. Leschot, and C. Heiting 1997, Genomics 39: 377-384;
Nelson, C.R., and P. Szauter 1992, Mol. Gen. Genet. 235: 11-21; Smith, P.A., and R.C. King 1968, Genetics
60: 335-351; Sym, M., and G.S.
Roeder 1995, J. Cell Biol. 128:
455-466; Tung, K.S., and G.S.
Roeder 1998, Genetics 149: 817-832;
Wayson, S.M., S.L. Page, B.W. Carey, M. Paddy, and R.S. Hawley 2001,
Abstracts of the 17th European Drosophila Research Conference: p.34;
Westergaard, M., and D. von Wettstein 1972, Ann. Rev. Genet. 6: 71-110; von Wettstein, D., S.W. Rasmussen, and P.B. Holm 1984, Ann.
Rev. Genet. 18: 331-413.