GadXW and Acid
Resistance Experimental Design
Experiment Summary and Publications
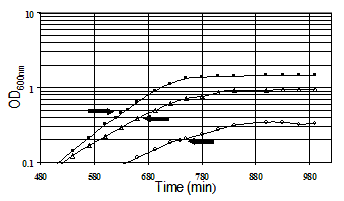
Growth of E. coli MG1655 (CGSC# 7740) growing at pH 7.2,
pH 5.5, and pH 4.5. Increasing acidity placed increasing burden
on growth. There was no difference in the growth curves of E.
coli DT162 (delta gadX::KanR), DT169 (delta gadXkan-),
DT203 (delta gadW::KanR), and DT232 (delta gadXgadW::KanR).
Arrows indicate the time of RNA isolation.
Culture Conditions. All cultures used for genomic expression profiling of acid resistance and GadXW regulon genes were grown in the minimal medium developed for E. coli proteome studies (Neidhardt et al., 1974). Glucose (0.2%) was the sole carbon and energy source. Morpholinepropanesulfonic acid (MOPS) was used as the buffer for pH 7.4 media and morpholinethanesulfonic acid (MES) was used to buffer the pH 4.5 and 5.5 media. Cultures were grown aerobically with 300 rpm agitation at 37 degrees C in 50 ml of medium in 250 ml fleakers. Growth was monitored by measuring the optical density (OD) at 600nm. RNA samples were isolated in mid-logarithmic phase by pipeting into ice-cold RNAlater™ (Ambion, Austin, TX) followed by purification using an RNeasy™ Mini Kit (Qiagen, Valencia, CA). The RNA samples were labeled by first strand cDNA synthesis. Labeled targets were hybridized to DNA arrays (Panorama E. coli Gene Arrays, Sigma Genosys Biotechnologies, Inc., The Woodlands, TX). The hybridized arrays were scanned by phosphorimaging at a pixel density of 100 microns (10,000 dots/cm2) with a STORM 820 PhosphoImager (Molecular Dynamics, Sunnyvale, CA) following exposure to a Kodak Storage Phosphor Screen GP (Eastman Kodak Co., Rochester, NY) for 24 hrs. The array membranes were consecutively hybridized, stripped, and rehybridized.
Spot-finding and quantitation. Image analysis software (ArrayVision, Imaging Research, Inc.) was used for spot-finding and quantitation of the E. coli Panorama arrays. The raw spot intensities were represented in a row-column format and exported into Microsoft Excel spreadsheets for further analysis, or as comma-delimited files (.csv) for upload to the database. Raw data from each experimental replicate were analyzed in Excel workbooks containing manually executed macros written in Visual Basic, or the data were processed in the database. The first step in the analysis associates the array coordinate for each spot with a unique spot number, the gene name, and related gene annotation. On the membrane arrays there are two spots for each gene, and these were treated as separate determinations. The raw data were normalized by expressing spot intensities as a percentage of the sum of all of the gene-specific spot intensities. The second step in the analysis applies the student t-test to determine the probability that the average of the experimental replicates is significantly different from the average of the control replicates . The P values (derived from the student t-test) for the normalized and natural log transformed data were calculated. The third step calculates relative gene expression between conditions by introducing a threshold value, chosen to be representative of the limit of detection of expressed genes (usually the 500th lowest expressed gene), and then calculating the ratio of the experimental/control expression levels such that genes that are more highly expressed in the experimental condition are given a positive value, and genes that are more highly expressed in the control condition are given a negative value.
Experiments. The goal of the gene expression profiling experiments was to identify acid-inducible target genes under GadX and or GadW control. The following gene expression profiles were obtained:
| Replicates |
||||
| Expt # |
Strain |
Condition |
Biological |
Technical |
| 1 |
Wt |
pH 7.4 |
2 |
5 |
| 2 |
Wt |
pH 7.4 |
1 |
2 |
| 3 |
Wt |
pH 5.5 |
2 |
5 |
| 4 |
Wt |
pH 5.5 |
1 |
2 |
| 5 |
Wt |
pH 4.5 |
1 |
2 |
| 6 |
gadX |
pH 7.4 |
2 |
5 |
| 7 |
gadX |
pH 7.4 |
1 |
2 |
| 8 |
gadX |
pH 5.5 |
2 |
5 |
| 9 |
gadX |
pH 5.5 |
1 |
2 |
| 10 |
gadX |
pH 4.5 |
1 |
2 |
| 11 |
gadX(Km-) |
pH 7.4 |
1 |
2 |
| 12 |
gadX(Km-) |
pH 5.5 |
1 |
2 |
| 13 |
gadW |
pH 7.4 |
1 |
2 |
| 14 |
gadW |
pH 5.5 |
1 |
2 |
| 15 |
gadX gadW |
pH 7.4 |
1 |
2 |
| 16 |
gadX gadW |
pH 5.5 |
1 |
2 |
Pair-wise Comparisons
Acid vs. Neutral Conditions
Exp 3 vs. 1; wildtype; pH 5.5 vs. pH 7.4
Exp 5 vs. 1; wildtype; pH 4.5 vs. pH 7.4
Exp 4 vs. 2; wildtype; pH 5.5 vs. pH 7.4
Exp 8 vs. 6; gadX; pH 5.5 vs. pH 7.4
Exp 10 vs. 6; gadX; pH 4.5 vs. pH 7.4
Exp 9 vs. 7; gadX; pH 5.5 vs. pH 7.4
Exp 12 vs. 11; gadX(kan-); pH 5.5 vs. pH 7.4
Exp 14 vs. 13; gadW; pH 5.5 vs. pH 7.4
Exp 16 vs. 15; gadXW; pH 5.5 vs. pH 7.4
Mutants vs. Wildtype under Neutral Conditions
Exp 6 vs. 1; pH 7.4; gadX vs. wildtype
Exp 7 vs. 2; pH 7.4; gadX vs. wildtype
Exp 11 vs. 2; pH 7.4; gadX(kan-) vs. wildtype
Exp 13 vs. 2; pH 7.4; gadW vs. wildtype
Exp 15 vs. 2; pH 7.4; gadXW vs. wildtype
Mutants vs. Wildtype under Acid Conditions
Exp 8 vs. 3; pH 5.5; gadX vs. wildtype
Exp 10 vs. 5; pH 4.5; gadX vs. wildtype
Exp 9 vs. 4; pH 5.5; gadX vs. wildtype
Exp 12 vs. 4; pH 5.5; gadX(kan-) vs. wildtype
Exp 14 vs. 4; pH 5.5; gadW vs. wildtype
Exp 16 vs. 4; pH 5.5; gadXW vs. wildtype
Data Legend
GadXW Data Set; Excel: 5.7 Mb)
1)WtpH7-4pct: average normalized spot intensity
for experiment 1; values expressed as a percentage of the sum of
all of the gene-specific spot intensities
3vs1)WtpH5-5vs7-4: log10 ratio of experiment 3
vs. experiment 1
3vs1)WtpH5-5vs7-4.p-ln: P value for the corresponding
log ratio calculated from the normalized, natural log transformed
data
E. coli Gene Expression Database (Oracle) Interface
Raw Data File Available on request