|
|
GadXW
Regulon and Glutamate-DependentAcid Resistance Ma, et al., 2002, J Bacteriol 184: 7001-7012 (PDF) Tucker, et al., 2003, J Bacteriol 185: 3190-3201 (PDF) Ma, et al., et al., 2003, Mol Microbiol 49: 1309-1320 (PDF) GadXW Experimental Design and Data Project Summary. E. coli MG1655 acid-inducible genes were identified by whole-genome expression profiling. Cultures were grown to mid-logarithmic phase on acidified glucose minimal medium, conditions that induce glutamate-dependent acid resistance (AR) while the other AR systems are either repressed or not induced. A total of 28 genes were induced in at least 2 of 3 experiments in which the gene expression profiles of cells grown in acid (pH 5.5 or 4.5) were compared to cells grown at pH 7.4. Several of these genes are clustered in the gadA region, including hdeA, which encodes a putative periplasmic acid chaperone, and 4 putative regulatory genes. One of these putative regulators, yhiE, was shown to significantly increase acid resistance when overexpressed in cells that had not been pre-induced by growth at pH 5.5 and mutation of yhiE decreased acid resistance; yhiE could therefore encode an activator of AR genes. Thus the acid-inducible genes clustered in the gadA region appear to be involved in glutatmate-dependent acid resistance, although their specific roles remain to be elucidated. GadX regulates two genes that encode isoforms of glutamate decarboxylase critical to this system, but additional genes associated with the glutamate-dependent acid resistance system remained to be identified. The gadX gene and a second downstream araC-like transcription factor, gadW, were mutated separately and in combination, and the gene expression profiles of the mutants were compared to the wild type strain grown in neutral and acidified medium under conditions favoring induction of glutamate-dependent acid resistance. Cluster and principal component analysis identified 15 GadX-regulated, acid-inducible genes. Reverse transcriptase mapping demonstrated that these genes are organized in 10 operons. Analysis of the strain lacking GadX but possessing GadW confirmed that GadX is a transcriptional activator under acidic growth conditions. Analysis of the strain lacking GadW but possessing GadX indicated that GadW exerts negative control over three GadX target genes. The strain lacking both GadX and GadW was defective in acid-induction of most but not all GadX target genes, consistent with the roles of GadW as an inhibitor of GadX-dependent activation of some genes and an activator of other genes. Resistance to acid was decreased under certain conditions in a gadX mutant and even more so by combined mutation of gadX and gadW. However, there was no defect in colonization of the streptomycin-treated mouse model by the gadX mutant in competition with the wildtype, and the gadX gadW mutant was a better colonizer than the wildtype. Thus, E. coli colonization of the mouse does not appear to require glutamate-dependent acid resistance. We favor a model of AR gene control in which GadX and GadW are intermediates in a regulatory cascade and serve to integrate signal(s) received by the cells to indicate they are present in an acid environment, have entered into stationary phase (RpoS), reflect medium composition (CRP), and additional unknown signals (HN-S and EvgA). This would leave the role of direct activation of target genes to one of the other transcription factors. In support of this hypothesis, we tested whether overproduction of YhiE could rescue AR in the gadX gadW mutant – it did (data not shown). Thus, we propose a complex regulatory cascade in which global regulators (RpoS, CRP, HN-S, EvgA, etc.) influence the expression levels and/or activities of GadX and GadW, which in turn activates the expression and/or activities of transcription activators (eg. YdeO and YhiE) that directly activate subsets of target genes involved in AR. One prediction of this model is that YhiE directly activates the glutamate-dependent AR genes. This cascade would allow the cell to integrate various physiological processes that are collectively important in AR. GadXW and Acid Resistance Experimental Design
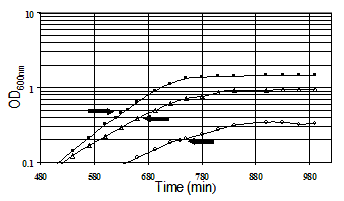
Culture Conditions. All cultures used for genomic expression profiling of acid resistance and GadXW regulon genes were grown in the minimal medium developed for E. coli proteome studies (Neidhardt et al., 1974). Glucose (0.2%) was the sole carbon and energy source. Morpholinepropanesulfonic acid (MOPS) was used as the buffer for pH 7.4 media and morpholinethanesulfonic acid (MES) was used to buffer the pH 4.5 and 5.5 media. Cultures were grown aerobically with 300 rpm agitation at 37 degrees C in 50 ml of medium in 250 ml fleakers. Growth was monitored by measuring the optical density (OD) at 600nm. RNA samples were isolated in mid-logarithmic phase by pipeting into ice-cold RNAlater™ (Ambion, Austin, TX) followed by purification using an RNeasy™ Mini Kit (Qiagen, Valencia, CA). The RNA samples were labeled by first strand cDNA synthesis. Labeled targets were hybridized to DNA arrays (Panorama E. coli Gene Arrays, Sigma Genosys Biotechnologies, Inc., The Woodlands, TX). The hybridized arrays were scanned by phosphorimaging at a pixel density of 100 microns (10,000 dots/cm2) with a STORM 820 PhosphoImager (Molecular Dynamics, Sunnyvale, CA) following exposure to a Kodak Storage Phosphor Screen GP (Eastman Kodak Co., Rochester, NY) for 24 hrs. The array membranes were consecutively hybridized, stripped, and rehybridized. Spot-finding and quantitation. Image analysis software (ArrayVision, Imaging Research, Inc.) was used for spot-finding and quantitation of the E. coli Panorama arrays. The raw spot intensities were represented in a row-column format and exported into Microsoft Excel spreadsheets for further analysis, or as comma-delimited files (.csv) for upload to the database. Raw data from each experimental replicate were analyzed in Excel workbooks containing manually executed macros written in Visual Basic, or the data were processed in the database. The first step in the analysis associates the array coordinate for each spot with a unique spot number, the gene name, and related gene annotation. On the membrane arrays there are two spots for each gene, and these were treated as separate determinations. The raw data were normalized by expressing spot intensities as a percentage of the sum of all of the gene-specific spot intensities. The second step in the analysis applies the student t-test to determine the probability that the average of the experimental replicates is significantly different from the average of the control replicates . The P values (derived from the student t-test) for the normalized and natural log transformed data were calculated. The third step calculates relative gene expression between conditions by introducing a threshold value, chosen to be representative of the limit of detection of expressed genes (usually the 500th lowest expressed gene), and then calculating the ratio of the experimental/control expression levels such that genes that are more highly expressed in the experimental condition are given a positive value, and genes that are more highly expressed in the control condition are given a negative value. Experiments. The goal of the gene expression profiling experiments was to identify acid-inducible target genes under GadX and or GadW control. The following gene expression profiles were obtained:
Pair-wise Comparisons Acid vs. Neutral Conditions
Data Legend GadXW
Data Set; Excel: 5.7 Mb) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
 |
|
Copyright © 2009 The Board of Regents of the University of Oklahoma | Disclaimer OU Bioinformatics Core Facility @ Advanced Center for Genome Technology | Credits | updated:19 Oct 2005 |