
Almeida, L.M., J.P. Castro, and C.M.A. Carareto. 2001. Micropia transposable element occurrence in Drosophila species of the saltans group. Dros. Inf. Serv. 84: 114-117.
|
|
|
|||
|
|
||||
Micropia transposable element occurrence in Drosophila species of the saltans group.
Almeida, L.M., J.P. Castro, and C.M.A. Carareto. Departamento de Biologia – IBILCE – UNESP. Rua Cristóvão Colombo 2265 CEP 15054-000, São José do Rio Preto – SP, Brasil; email: carareto@bio.ibilce.unesp.br
Introduction
Transposable
elements origin and distribution in different species and whether they are
recent or old components of the host genome are still subjects of research.
The presence of a transposable element in all organisms of a species
group suggests that they are old components of these genomes.
Members of the retrotransposon family micropia were discovered as constituents of wild-type Y chromosomal
fertility genes from Drosophila hydei (Hennig et al., 1983; Huijser et
al., 1988), but they also occur in autosomes
and X chromosomes. The presence
of the micropia retrotransposon
in different Drosophila species was determined by Southern analysis using the micropia element of D. hydei as a probe. It
has been found in several species of the all subgroups of the repleta group, but with patchy distribution (Lankenau et
al., 1994); in the immigrans group, in D. immigrans, in the funebris group, in D. funebris, and in the mellanica group, in D. mellanica (Lankenau, 1993); and in three of the four species
groups of the subgenus Sophophora: melanogaster group (D.
melanogaster, D. simulans,
D. birchii, D.
yakuba, D. ananassae); willistoni group (D.
willistoni); and the saltans group (D. saltans) (Lankenau and Hennig, 1990; Lankenau et al., 1988; Lankenau
et al., 1990;
Lankenau et al., 1994). Micropia
has been studied in details only in species of the repleta group, D. hydei in special,
and in D. melanogaster. It has a typical retrotransposon structure
with approximately 5.5 kb length and a 4 kb open reading frame encoding putative
products that show homology to the nucleocapsid, protease, reverse transcriptase,
RNase H and integrase products of vertebrate retroviruses (Lankenau et
al., 1988;
Lankenau et al., 1989). The
general structure of this retrotransposon is well conserved in both species,
but the LTRs are completely different. The overall sequence homology ranges between 70% and 90% on
the amino acid level. Micropia encodes a 5.0 kb transcript that is expressed in both
testes and somatic tissues of males and females. Although the great similarity,
micropia in D. hydei
produces an antisense RNA overlapping the RNaseH and parts of the transcriptase
reverse that is not expressed in D. melanogaster. Since only D. saltans was screened for the presence of micropia in its genome, this study aimed to contribute to the knowledge about micropia
distribution in the saltans
species group.
Material and Methods
Species: The species of D. saltans
group studied are listed in Table 1.
PCR reactions:
PCR reactions were performed in 25ml volumes
using approximately 200ng of template DNA, 100 mM of
each dNTP, 12 pmol of each primer, 1.5 mM of MgCl2 and 1 unit of Taq DNA Polymerase (GIBCO-BRL) in 1´
Polymerase Buffer. After an initial
denaturation for 3 min at 95°C, 40 cycles consisting of a
1-min denaturation at 95 °C, a 1-min annealing at 52°C
and a 2-min extension at 72°C steps were followed. An additional extension step of 10 min at 72°C
was performed after the last cycle.
The amplified fragments were separated by electrophoresis in a 1% agarose
gel. The primers used were #2813
(5’- TTAACTCCTAGAGTTCATCGCTGG- 3’) and #2814 (5’- CATGTACCTGGTTAACTACTGACC
- 3’) which amplify a 386 bp fragment from a highly conserved sequence
of micropia (from nucleotide 2813 to
3198).
Dot blot hybridization:
Denaturated DNA (5 mg) was applied directly
to the nylon membrane and hybridized with a 3.1 kb micropia fragment excised with EcoRI from
dhMiF2 plasmid (Huijser et al., 1988). Hybridization and
detection were performed with ECLTM direct nucleic acid labeling
and detection systems according to manufacturer’s instructions.
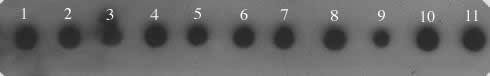
Figure 1. Dot blot
of D. saltans species group probed with a 3.1 kb micropia
fragment. The genomic DNAs were blotted as follow: 1. D. neocordata; 2. D. emarginata; 3. D. parasaltans; 4. D. subsaltans; 5. D. milleri; 6. D. dacunhai; 7. D. sturtevanti; 8. D. austrosaltans; 9. D. saltans; 10. D. prosaltans; 11. micropia (dhMiF2 plasmid).
Results and Discussion
|
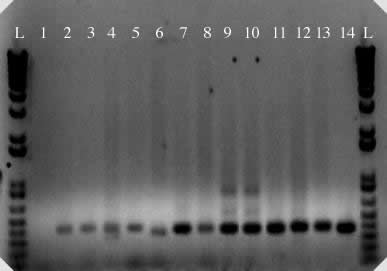
Figure 2. PCR
amplification of micropia internal sequence from D. saltans
species group and control DNAs. 1. negative control; 2. D. neocordata; 3. D. emarginata; 4. D. parasaltans; 5. D. subsaltans; 6. D. milleri; 7. D. dacunhai; 8. D. sturtevanti; 9. D. austrosaltans; 10. D. saltans;
11. D. prosaltans;
12. D. simulans;
13. D. melanogaster; 14. micropia (dhMiF2 plasmid). L = 1 kb plus DNA ladder.
We here present a contribution to the knowledge of the distribution of micropia retrotransposon in the saltans group species. According to literature, this is the first description of micropia occurrence in species of the subgroups cordata (
D. neocordata), elliptica (D. emarginata), sturtevanti (D. milleri, D. dacunhai, D. sturtevanti), and parasaltans (D. parasaltans and D. subsaltans,) and in other species of the saltans subgroup than D. saltans (D. austrosaltans and D. prosaltans). Since the saltans group belongs to the Sophophora subgenus, additional analyses are in progress in our laboratory aiming to determine whether the antisense RNA has still been transcribed in these species, or not, as in D. melanogater, a species closer to saltans than to repleta species group.
Acknowledgments: We thank D-H. Lankenau (German Cancer Research Center, Heidelberg, Germany) for providing us with dhMiF2 plasmid. This research was supported by grants and fellowships of FAPESP and CNPq.
References: Biémont, C., and G. Cizeron 1999, Genetica 105: 43-62; Hennig W., P. Huijser, P. Vogt, H. Jäckle, and J-E. Edström 1983, EMBO J. 2: 1741-1746; Huijser, P., C. Kirchhoff, D-H. Lankenau, and W. Hennig 1988, J. Mol. Biol. 203: 689-697; Lankenau, D-H., 1993, In: Transposable Elements and Evolution. (McDonald, J., ed.). pp. 232-241. Kluwer, Dordrecht, The Netherlands; Lankenau, D-H., and W. Hennig 1990, Nucl. Acids Res. 18: 4265-4266; Lankenau, D-H., P. Huijser, E. Jansen, K. Miedema, and W. Hennig 1988, J. Mol. Biol. 204: 233-246; Lankenau, D-H., P. Huijser, W. Hennig 1989, J. Mol. Biol. 209: 493-497; Lankenau, D-H., P. Huijser, E. Jansen, K. Miedema, and W. Hennig 1990, Chromosoma 99: 111-117; Lankenau, S., V.G. Corces, and D-H. Lankenau 1994, Mol. Cell. Biol. 14: 1764-1775.